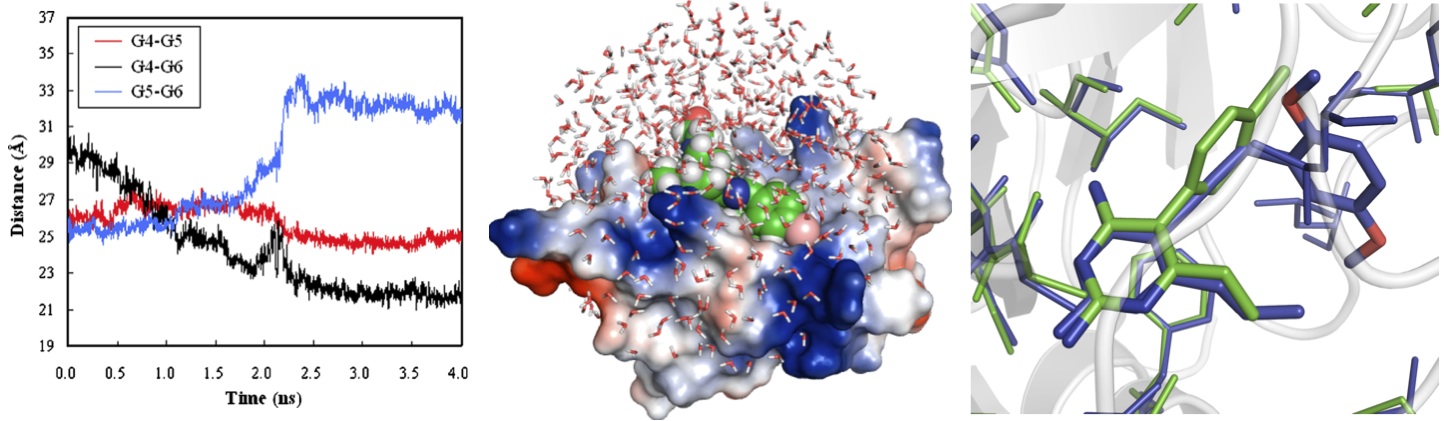
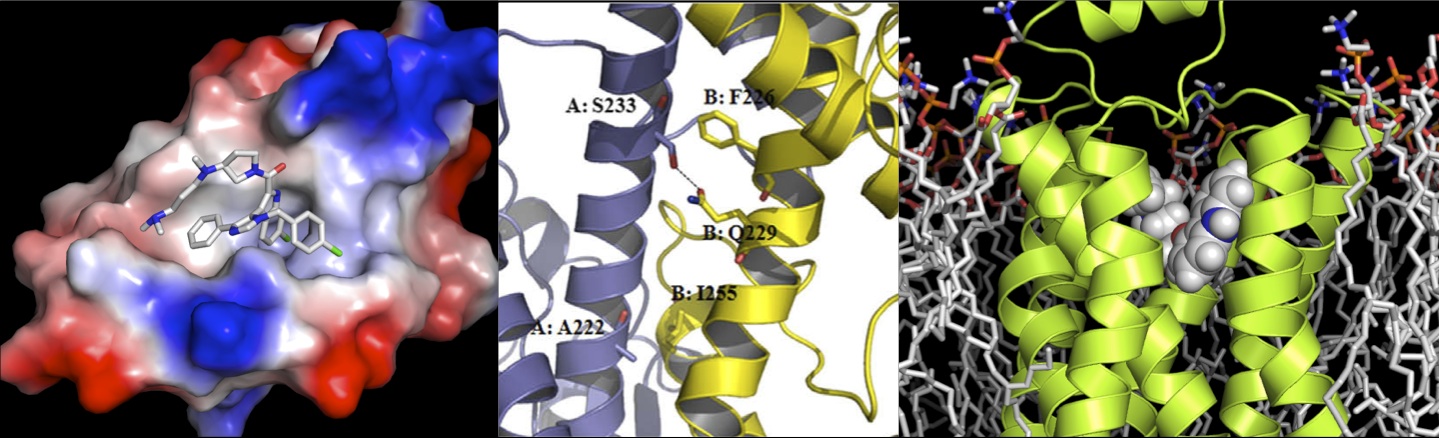
Structural Systems Pharmacology
My research works have centered on Molecular Recognition in Biological Systems by harnessing computational and theoretical approaches. To that end, I have been pursuing to make research achievements based on expertise on multi-scale computational modeling approaches, ranging from atomistic simulation and molecular docking to high throughput virtual screening and proteomic scale structural analysis.
How molecular recognition can be quantified and optimized is the most important issue in drug discovery field.
Computer Aided Drug Design (CADD) is a specialized field that uses computational methods to speed up the drug development process.
The ultimate aim of my research is to develop an advanced computational platform for “interactome” study in the context of the three dimensional structures of biomolecules to obtain fundamental understanding of the molecular mechanisms of human diseases and to apply it to rational CADD.
>> My research interests
about
Hui Sun Lee
Assistant Research Professor
Higuchi Biosciences Center
The University of Kansas
ContacT information
Center for Computational Biology
Department of Molecular Biosciences
The University of Kansas
2030 Becker Drive,
Lawrence, KS 66045
Office phone: (785)864-1966
E-mail : huisun.cadd@gmail.com
Recent publications
L-Met Activates Arabidopsis GLR Ca2+ Channels Upstream of ROS and Regulates Stomatal Movement.
D. Kong, H-C Hu, E. Okuma, Y. Lee, H. S. Lee, S. Munemasa, D. Cho, L. Pedoeim, B. Rodriguez, C. Ju, W. Im, Y. Murara, Z-M. Pei, and J. M. Kwak. (submitted)
G-LoSA: An Efficient Computational Tool for Local Structure-Centric Biological Studies and Drug Design.
H.S. Lee and W. Im, Protein Sci. 25: 865-876 (2016)
Converting One-Face α-Helix Mimetics into Amphiphilic α-Helix Mimetics as Potent Inhibitors of Protein-Protein Interactions.
J.H. Lee, M. Oh, H.S. Kim, H.S. Lee, W. Im, and H-S. Lim, ACS Comb. Sci. 18: 36-42 (2016)