G-LoSA (Graph-based Local Structure Alignment) is an efficient computational method to align protein local structures in a sequence order independent way and to provide the GA-score (between 0 and 1), a size-independent quantity of structural similarity for a given local structure pair.
In particular, the GA-score is calculated based on the chemical features of each amino acid and can be applied to measure the structural similarity with the local structures of diverse sizes and characteristics, yet maintaining its length independency.
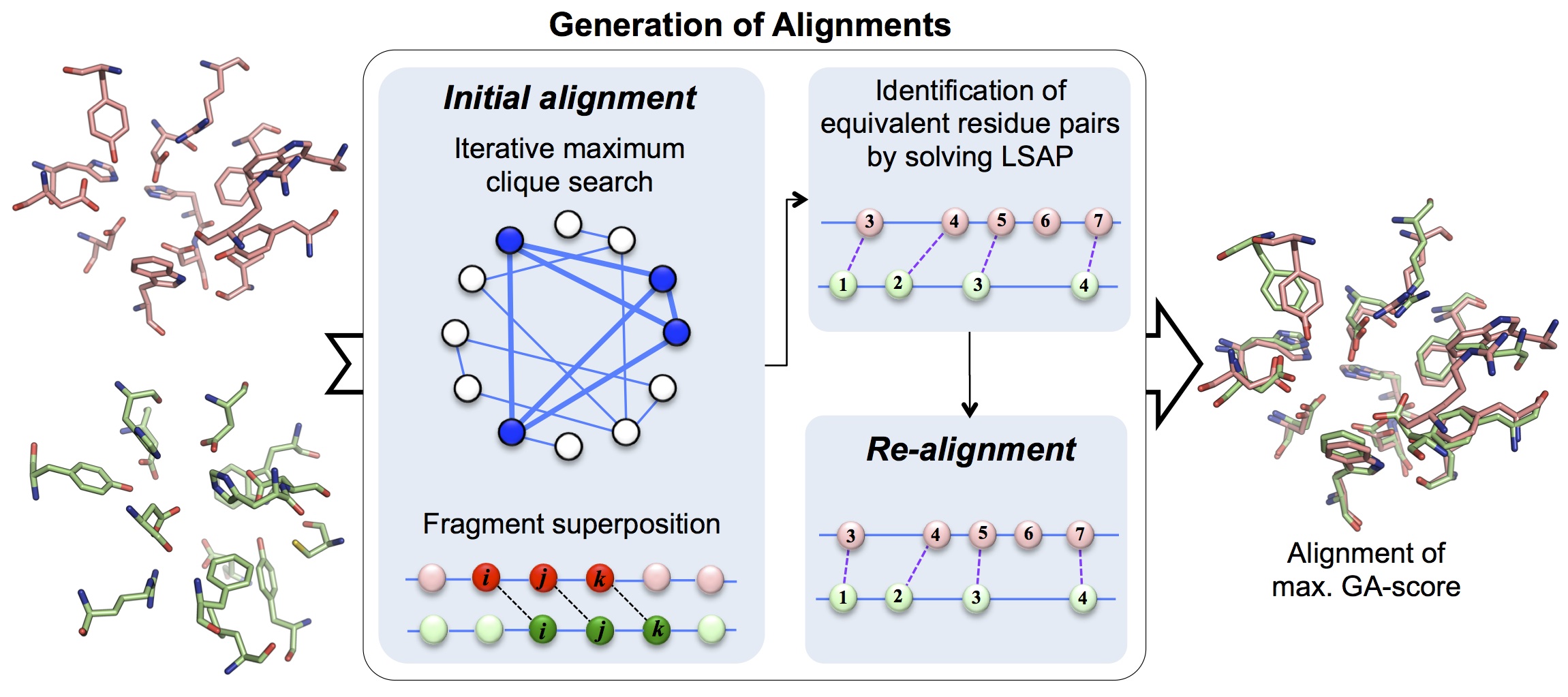
In G-LoSA, both the iterative maximum clique search and the fragment superposition are adopted to generate all possible alignments between two different local structures and the optimal alignment is determined by the maximum GA-score (figure below).
1. Installation
|
Click here to download the G-LoSA software package including example PDB local structures.
The source code of G-LoSA (glosa.cpp) is written in C++ and can be easily compiled using C++ GNU compiler by |
|
|
>g++ -c glosa.cpp >g++ -o glosa glosa.o |
2. Preparing Chemical Feature and Secondary Structure Files
|
Input protein structure files for G-LoSA must be in PDB format and be ended with "TER". |
|
|
G-LoSA needs chemical feature file for each input protein structure to calculate GA-score. Users can also optionally use a secondary structure file for each input protein structure to reduce computational time. The G-LoSA software package contains auxiliary programs (AssignChemicalFeatures.java and AssignSecondaryStructures.java) to easily generate these chemical feature and secondary structure files. |
|
|
* For chemical feature information
* For secondary structure information |
3. Running G-LoSA
| -s1 | structure 1 (PDB format) | |
| -s1cf | chemical feature file for structure 1 | |
| -s2 | structure 2 (PDB format) | |
| -s2cf | chemical feature file for structure 2 | |
| [advanced options] | ||
| |
4. Outputs
GA-score |
G-LoSA alignment score |
|
ali_struct.pdb |
PDB coordinates of s2 structure aligned onto s1 structure |
|
matrix.txt |
translational and rotational matrix to align -s2 structure onto -s1 structure |
|
ali_struct_with.pdb |
PDB coordinates of s2w structure transferred by the matrix |
|
| |
G-LoSA Toolkit
| Please visit here for G-LoSA Toolkit. |
|
References
| ||
| |
Note
| ||
| |
G-LoSA Update History
* For any question or suggestion, e-mail to Hui Sun Lee (huisun.cadd@gmail.com)
